Part 1. The DNA Sequencing Arms Race: The Fight to Dethrone Illumina
Multi-part series exploring the tech and politics around short/long-read sequencing advances
Humanity’s genomic literacy is advancing at an exciting pace. If you haven’t been paying attention to DNA sequencing tech, I suggest you tune in.
There's something marvellous (and also occasionally existentially destablizing) about the fact that the microscopic universe packed inside our bodies, the incredibly detailed genomic library neatly coiled in double-helix form within our cells, can be read back to us.
We have the capacity to decode ancient language encoded into our very beings. DNA also just happens to be the universal language encoded within all living things. This means that the technologies we use to read about ourselves can also be used to read the genome of organisms like bioluminescent dinoflagellates.
Now, for the past 15+ years, short-read sequencing (SRS), dominated by Illumina’s Next-Generation Sequencing platforms, has been the workhorse driving our ability to sequence (read) DNA. It essentially works by breaking DNA into 150 base pair fragments and reads them in parallel at massive scale. SRS made genome sequencing fast, high-throughput, and (relatively) affordable. Illumina’s short-read sequencing-by-synthesis (SBS) chemistry became the gold standard, giving the company over 80% market share in sequencing instruments and consumables.
Basically if you have been sequencing DNA, you were probably using Illumina.
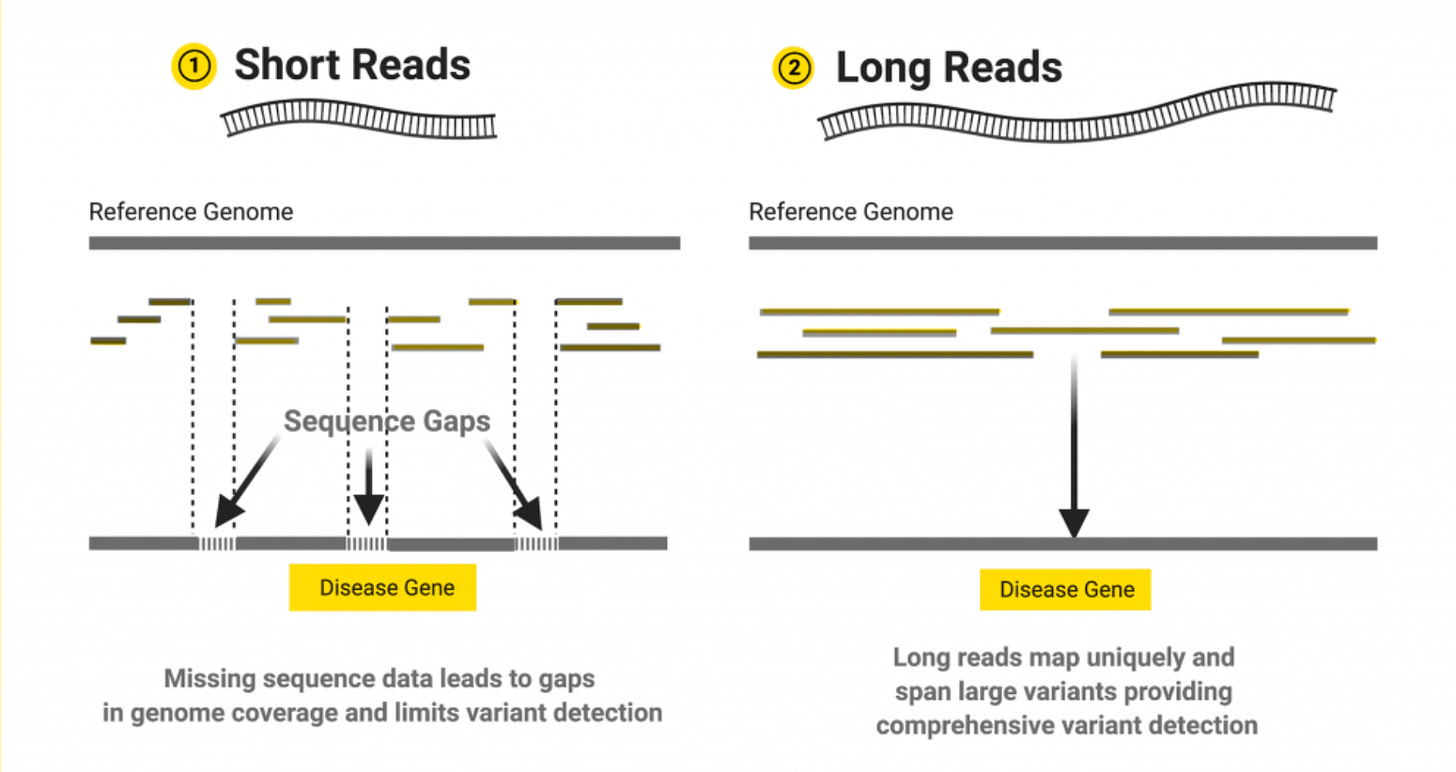
The Limitations of Short Read Sequencing
Even as Illumina’s SRS transformed genomics, its inherent limitations have become increasingly clear. For example, short reads struggle to map thing like complex genomic terrain, large structural variants, and long tandem repetitive regions. Highly similar sequences (like pseudogenes) confuse short-read alignment and critical information beyond the plain sequence itself are mostly lost on Illumina’s platform, which requires chemical tricks to detect modifications indirectly.
The short-read sequencing approach is kind of like trying to understand War and Peace by reading random 150-character tweets. Sure, if you try hard enough, you can get quite a bit of the critical plot points, but entire narrative arcs? Epic character developments? These can be lost in translation.
The obstacles in the short-read space have motivated researchers to seek better, longer-read technologies that can span kilobases or more. (side note: Microsoft and University of Washington researchers actually encoded War and Peace into DNA)
Brief Look at The Challengers: SBX, Nanopore, HiFi Long Reads and China
Over the last few years, we’ve seen the rise of several interesting challengers to Illumina’s short-read monopoly and the inherent limitations of the tech:
Sequencing by Expansion (SBX), a new approach spearheaded by Roche’s acquisition of Stratos Genomics is saying bigger doesn’t necessarily mean better. They are promising to boost short-read accuracy and sequencing speed substantially.
Oxford Nanopore Technologies (ONT) has popularized real-time, long-read sequencing, where ultra-long reads and direct DNA/RNA analysis (including native methylation) offer a qualitatively different view of the genome as well as portable options
PacBio’s HiFi (High-Fidelity) long reads now routinely reach >15 kb read lengths with Q30-Q40 accuracy (~99.9%), delivering highly accurate assemblies that reveal variants that short reads just miss.
All the while, a geopolitical undercurrent is shaking up the landscape as China’s highly prioritized biotechnology industry grows quickly. In response to US tariffs, China has even blacklisted Illumina as an “unreliable entity”. China continues to cultivate domestic champions like BGI Group and its instrument arm MGI Tech. As Chinese sequencers begin competing globally (most certainly at lower price points), we will see the once Illumina centred genome market fracture and fragment along these next gen. fault lines.
This is part 1 of a series where I’ll analyze the sequencing landscape, profile the key challengers in short and long read sequencing (SBX, nanopore, HiFi long reads), explore why the shift away from short-read tech is happening now, examine China’s growing role, and assess who might win the sequencing race in the next decade.
The genomics field is at an inflection point where technical innovation, market economics, and even international politics are redefining what the next generation of sequencing will look like. For market share giant Illumina, the future may be filled with less certainty and interestingly, more competition.
Why the Shift is Happening Now
Illumina’s short-read SBS chemistry has been refined to astonishing levels of accuracy and throughput, with the latest NovaSeq X machines capable of doing up to 26 billion single reads per flow cell. So why are researchers increasingly looking beyond short reads?
The fundamental issue is ultimately that you can’t assemble what you can’t read. No matter how high you pile up 150 bp reads, contiguous structures larger than that remain black boxes. Roughly 5–10% of the human genome (much of it repetitive or structurally complex) is considered “dark” to Illumina sequencing, meaning important variants in these regions (which could underlie rare genetic diseases) are missed by default. Short reads also suffer biases like extreme GC content regions may sequence poorly and they typically require PCR amplification, which can introduce errors or drop out regions and also critically wipes out native epigenetic markings.
Long reads offer a way to leap over those hurdles by simply reading DNA in bigger chunks. But until recently, long-read methods came with a tradeoff of much higher raw error rates and cost.
Critically, in the last few years, long-read accuracy and throughput have improved greatly while costs continue to fall, making them way more practical. PacBio’s introduction of HiFi reads (through circular consensus sequencing) made the major move to prove that you can have both long read length and short read accuracy in one technology.
PacBio compared their new Revio system to Illumina’s SBS and Nanopore sequencing technology in the table below:
Oxford Nanopore’s continuous upgrades to its pore chemistry and basecalling algorithms for new kits have pushed raw read accuracies into the 99% range and even higher for duplex reads.
Meanwhile, short-read costs are having diminishing returns, the famed $1,000 genome is now well below $1,000 in consumables on high-end instruments. In fact, today a human genome can be sequenced for as little as $200 in consumables on Illumina’s NovaSeq X or on newcomer short-read platforms like Element Biosciences’ Aviti system. Ultima Genomics has even demonstrated a $100 genome in early access.
With such low costs, sequencing volume is exploding and new applications in clinical diagnostics, direct-to-consumer genome scans, population sequencing projects are all coming online. However, application like these once again bring up the questions around “dark” genomic matter and all of the information that we’re not getting. How critical might that be for disease and areas like personalized medicine. This realization is driving uptake of long-read and specialty sequencing for cases where short reads literally fall short.
Another factor is the error-correction problem: Certain genomic variants can mimic sequencing errors and vice-versa. For example, distinguishing a true 10-base insertion in a repeat region from a sequencing noise artifact is extremely difficult with short reads, you either get lots of false positives if you loosen criteria, or miss real variants if you tighten.
Long reads with higher accuracy solve this by physically covering the entire variant in one read, leaving little ambiguity. In cancer, short reads might mis-sequence a hyper-mutated segment and incorrectly call mutations that aren’t really there or miss ones that are. High-fidelity long reads greatly reduce these false calls. In other words, longer reads can act as their own error-correction, because each read gives more context.
Market and Regulatory Landscape
Market economics are also driving the shift. For years, Illumina short reads were so much cheaper and better than any competitors that sticking with Illumina was a no-brainer for most labs. That math is no longer ‘math-ing’ in quite the same way for all use cases.
If in the not too far future you can get 90% of the information from a genome with short reads for $100, but 100% of the information, including structural variants, phasing, epigenetics, with long reads for 5x or 10x the cost and quickly, researchers and clinicians might consider the extra cost worth it. The value of a missed diagnosis or an undiscovered structural variant can be much higher than the difference in sequencing cost.
One compelling statistic states that in rare genetic disease diagnostics, short-read genome sequencing fails to find a cause in roughly 50% of cases (which feels insanely high). Adding long-read sequencing can yield a diagnosis in an additional 10-20% of those cases (i.e. catching structural variants or repeat expansions that Illumina missed). Success stories like these are pushing hospitals to pilot long-read sequencing for neurodevelopmental disorders, muscular dystrophies, and more, especially as costs go down.
On the regulatory and clinical adoption side, we’re at an early but pivotal stage. Regulatory agencies have begun to approve or validate genomic tests using long read sequencing, and there’s movement toward standardizing long-read sequencing for certain diagnostics.
For example in 2024, Azenta became the first commercial provider to obtain regulatory approval to offer clinical long-read sequencing whole genome sequencing tests with PacBio’s Revio sequencer in the USA.
So, are short reads out?
All of that being said, short reads are far from obsolete, they remain the backbone of most sequencing done today and will coexist with new methods for years to come. The SRS market is projected to grow substantially and the dominant short read blind spots have only opened the sequencing door wider for smarter, deeper and more innovative explorations in both the short and long read space.
Perhaps the more interesting question isn’t whether short reads will lose dominance, but rather aside from the clear use cases, what are we going to do with all this new high quality data we’re now able to generate?
Our technologies often outpace our frameworks for understanding. In these genomic data sets, we are being handed answers to questions we haven’t figured out how to ask yet. We’re sequencing more than ever, but interpreting what we find is still messy, fragmented, and still an emerging frontier.
The key here is finding the balance between extracting more data and extracting more meaning.






I am waiting :) https://thewinterbutton.substack.com/p/rain-translation-system-by-2028
A brilliant analysis (as per usual) of the genomics landscape. I’m very excited about the capacity for Chinese innovation to increase the availability of NGS tech globally. Of course the mission for finding meaning is equally critical, there are quite a few plug and play genome analytical services out there in SaaS companies or open source entities like Galaxy, but QC especially on an organismal idiosyncratic level is properly a future hurdle.